Background: Tumor development is intricately influenced by metabolic reprogramming, resulting in the emergence of exploitable vulnerabilities that can be targeted. Glutamine (Gln) depletion has emerged as a promising therapeutic strategy for malignancies, including AML, that heavily rely on Gln for their survival. Erwinia-derived asparaginase, also called crisantaspase, has exhibited anti-AML activity in both preclinical and clinical models by disrupting Gln supply, albeit with the persistence of residual leukemia burden. To identify cellular pathways modulated after crisantaspase-induced Gln depletion, here we report our comprehensive results using a whole transcriptome profiling approach (RNA-Seq) after crisantaspase treatment.
Methods: Whole-transcriptomic profiling using RNA-Seq was performed to profile gene expression of MOLM-14 AML cells following crisantaspase treatment. RNA was isolated from each sample and total RNA was utilized for this study. The samples were sequenced at IGS using the Illumina HiSeq sequencing platform. Raw sequencing reads generated for each sample were analyzed using the CAVERN analysis pipeline. Read quality was assessed using the FastQC toolkit to ensure good-quality reads for downstream analyses. P values were generated using a Wald test implemented in DESeq2 and then corrected for multiple hypothesis testing using the Benjamini-Hochberg correction method.
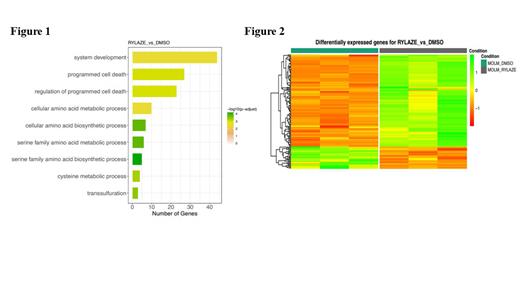
Results: We utilized Rylaze, an FDA-approved Erwinia crisantaspase, to deplete exogenous Gln. MOLM-14 cells were treated with Rylaze for 16h, followed by whole-transcriptome/gene expression profiling (GEP) using RNA-seq. The transcriptomic analysis provided information on the regulation of gene expression between vehicle control and Rylaze treatment in AML, as well as on the potential association with several putative pathways including serine family amino acids biosynthetic process (GO: 0009070, p-value = 2.18E-08), regulation of programmed cell death (GO:0043067, p-value = 7.96E-06), and amino acid transmembrane transport (GO:0003333, p-value = 5.10E-05). Furthermore, Figure 1 illustrates the gene ontology terms enriched by the DEGs, offering insights into the underlying molecular mechanisms involved in the response to Rylaze-induced Gln depletion in MOLM-14 cells. For example, the upregulation of genes associated with the serine family amino acid biosynthetic process may suggest a compensatory mechanism to counteract the depletion of Gln induced by Rylaze. By employing generalized linear regression models, we observed significant changes in gene expression following Rylaze (141 genes) compared to the vehicle control. This finding is further illustrated by a heatmap generated from transcriptome data analysis (Figure 2). We identified 141 significantly differentially expressed genes (DEGs); among the Rylaze-treated samples, consisting of 114 up-regulated and 27 down-regulated genes. Specifically, we focused on five genes that exhibited significant modulation in response to the treatment with Rylaze. These genes included CHAC1, ADM2, S100P, CTH and SPOCK2. These genes might play crucial roles in the cellular response to Gln depletion and could potentially serve as biomarkers or therapeutic targets in AML treatment. Validation of these observations was performed using immunoblotting techniques and quantitative real-time polymerase chain reaction (qRT-PCR), which confirmed the significant increase in CHAC1, ADM2, S100P, and CTH, as well as the significant decrease in SPOCK2 (p<0.0001) following Rylaze-induced Gln depletion. These findings contribute to a better understanding of the drug's mode of action and the impact of Gln depletion on cellular processes in AML.
Conclusion: This study represents a significant contribution towards advancing our understanding of amino acid-regulated metabolic pathways in AML, with potential implications for other Gln metabolism-dependent cancers as well.
OffLabel Disclosure:
Emadi:Kite Pharma: Consultancy, Membership on an entity's Board of Directors or advisory committees; Servier: Research Funding; Amgen: Membership on an entity's Board of Directors or advisory committees, Research Funding; NewLink Genetics: Research Funding; Jazz Pharmaceuticals: Research Funding; Genentech: Membership on an entity's Board of Directors or advisory committees; Secura Bio: Consultancy; KinaRx, Inc: Membership on an entity's Board of Directors or advisory committees, Other: Co-founder.
RYLAZE (asparaginase erwinia chrysanthemi- recombinant-rywn) is indicated as a component of a multi-agent chemotherapeutic regimen given by intramuscular injection for the treatment of acute lymphoblastic leukemia (ALL) and lymphoblastic lymphoma (LBL) in adult and pediatric patients 1 month or older who have developed hypersensitivity to E. coli-derived asparaginase.

This feature is available to Subscribers Only
Sign In or Create an Account Close Modal